Modeling
biological processes are complicated
Model types
A mathematical model is a representation of the essential aspects of a system [...] which presents knowledge of that system in usable form.
Pieter Eykhoff 1974
All models are wrong, but some are useful.
George Box 1976
The inverse problem
Parameter inference
Basic idea
Basic idea
e.g. an ODE model:
states: $\dot x (t, \theta) = f(t, x, \theta)$, $x(t_0, \theta) = x_0(\theta) \in \mathbb{R}^{n_x}$,
observables: $y(t, \theta) = h(x(t, \theta), \theta) \in \mathbb{R}^{n_y}$,
measurements: $\bar y_{ik} \sim \mathcal{N}(y_k(t_i,\theta), \sigma^2_{ik}(\theta) \in \mathbb{R}^{n_y}$
likelihood function: $p(D|\theta) = \prod_{ik}\frac{1}{\sqrt{2\pi}\sigma_{ik}(\theta)} \exp\left[-\frac{1}{2} \left(\frac{\bar y_{ik} - y_k(t_i,\theta)}{\sigma_{ik}(\theta)}\right)^2\right]$
objective function: $J(\theta) := - \log p(D|\theta) = \frac{1}{2}\sum_{ik}\left[\log(2\pi\sigma_{ik}^2(\theta)) + \left(\frac{\bar y_{ik} - y_k(t_i,\theta)}{\sigma_{ik}(\theta)}\right)^2\right] \overset{!}{\rightarrow}\min_{\theta\in\Theta}$
if prior knowledge is available: prior $p(\theta)$,
$J(\theta) := -\log p(D|\theta) - \log p(\theta)$
states: $\dot x (t, \theta) = f(t, x, \theta)$, $x(t_0, \theta) = x_0(\theta) \in \mathbb{R}^{n_x}$,
observables: $y(t, \theta) = h(x(t, \theta), \theta) \in \mathbb{R}^{n_y}$,
measurements: $\bar y_{ik} \sim \mathcal{N}(y_k(t_i,\theta), \sigma^2_{ik}(\theta) \in \mathbb{R}^{n_y}$
likelihood function: $p(D|\theta) = \prod_{ik}\frac{1}{\sqrt{2\pi}\sigma_{ik}(\theta)} \exp\left[-\frac{1}{2} \left(\frac{\bar y_{ik} - y_k(t_i,\theta)}{\sigma_{ik}(\theta)}\right)^2\right]$
objective function: $J(\theta) := - \log p(D|\theta) = \frac{1}{2}\sum_{ik}\left[\log(2\pi\sigma_{ik}^2(\theta)) + \left(\frac{\bar y_{ik} - y_k(t_i,\theta)}{\sigma_{ik}(\theta)}\right)^2\right] \overset{!}{\rightarrow}\min_{\theta\in\Theta}$
if prior knowledge is available: prior $p(\theta)$,
$J(\theta) := -\log p(D|\theta) - \log p(\theta)$
Optimization
find the best point estimate (MLE)- local vs global or multistart
- derivative-free vs derivative-based
- higher-order information
Hierarchical optimization
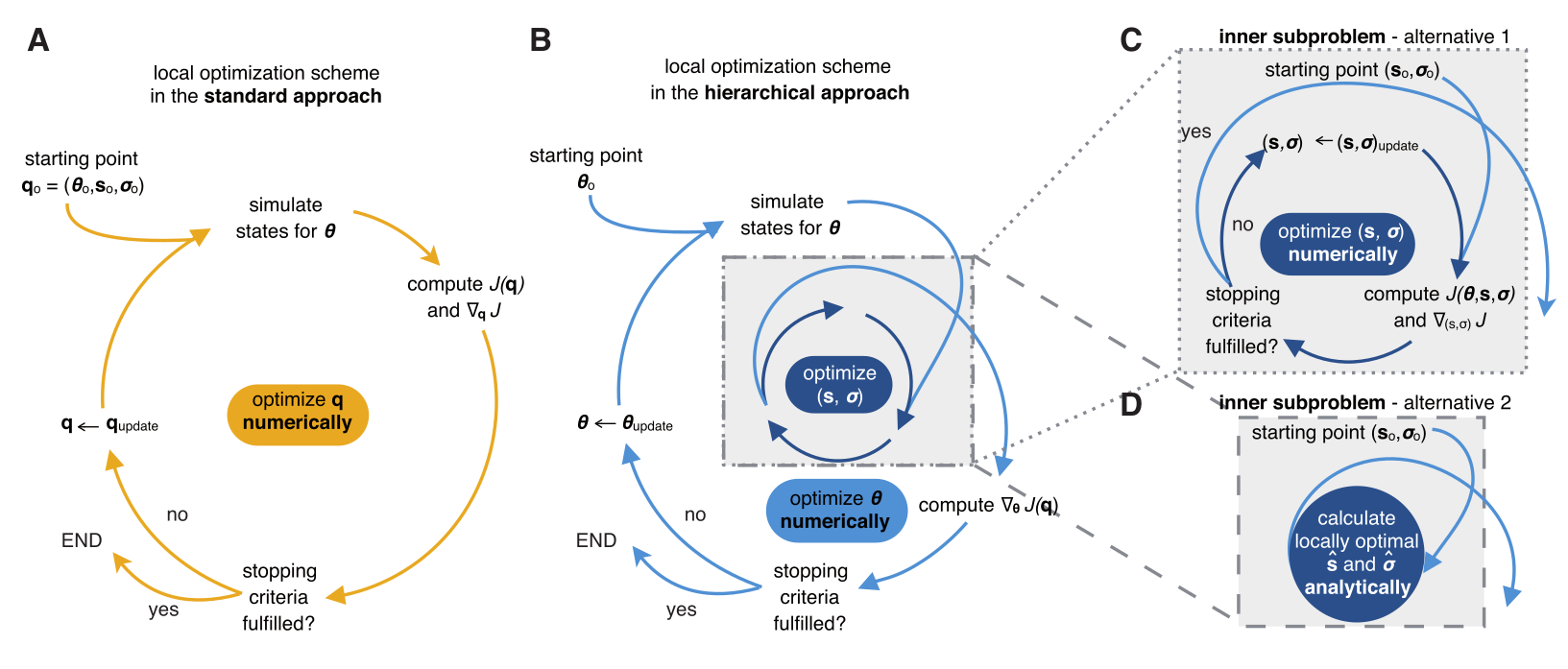 Loos, Carolin, et al.
"Hierarchical optimization for the efficient parametrization
of ODE models." Bioinformatics 2018.
Loos, Carolin, et al.
"Hierarchical optimization for the efficient parametrization
of ODE models." Bioinformatics 2018.
Computing gradients $\nabla J(\theta)$
$$\nabla J(\theta) = \sum_{i=1}^{n_t}\left[\frac{\partial J_i}{\partial y}(y(t_i,\theta),\theta)\color{red}{\frac{dy}{d\theta}(t_i,\theta)} + \frac{\partial J_i}{\partial \theta}(y(t_i,\theta),\theta)\right]$$- finite differences: $\delta_l J(\theta) = (J(\theta + \delta e_l) - J(\theta)) / \delta$
- forward sensitivities: calculate $\frac{dx}{d\theta}$ via additional ODEs
- adjoint sensitivities: circumvent $\frac{dx}{d\theta}$ via a cheaper adjoint state
finite differences
$$\delta^{h}_lJ(\theta) = \frac{J(\theta + \frac 1 2h\cdot e_l) - J(\theta - \frac 1 2h\cdot e_l)}{h}$$- inefficient: $O(n_xn_\theta)$
- numerically unstable
forward sensitivities
$$\nabla J(\theta) = \sum_{i=1}^{n_t}\left[\frac{\partial J_i}{\partial
y}(y(t_i,\theta),\theta)\frac{dy}{d\theta}(t_i,\theta) + \frac{\partial
J_i}{\partial \theta}(y(t_i,\theta),\theta)\right]$$
where, with $s^x = \frac{dx}{d\theta}$, the extended ODE
$\dot x = f, x(0) = x_0$,
$\dot s^x_l = \frac{\partial f}{\partial x}s^x_l + \frac{\partial f}{\partial \theta_l}, s^x_l(0) = \frac{\partial x_0}{\partial\theta_l}, l=1,\ldots,n_\theta$
allows us to calculate $\frac{dy}{d\theta}$
$\dot x = f, x(0) = x_0$,
$\dot s^x_l = \frac{\partial f}{\partial x}s^x_l + \frac{\partial f}{\partial \theta_l}, s^x_l(0) = \frac{\partial x_0}{\partial\theta_l}, l=1,\ldots,n_\theta$
allows us to calculate $\frac{dy}{d\theta}$
- inefficient: $O(n_xn_\theta)$
- numerically robust
- can exploit ODE structure
adjoint sensitivities
Idea: introduce an adjoint state $p\in\mathbb{R}^{n_x}$ such that $$\nabla J(\theta) = \sum_{i=1}^{n_t}\left[\frac{\partial J_i}{\partial\theta} + \frac{\partial J_i}{\partial y}\frac{\partial h}{\partial\theta}\right] - p(t_0)^T\frac{d x_0}{d\theta}(\theta) - \int_{t_0}^{t_{n_t}}p^T\frac{\partial f}{\partial\theta}d\tau$$ without the annoying $\frac{dx}{d\theta}$!$p$ is uniquely defined by the backward ODE $$\dot p = -\frac{\partial f}{\partial x}^Tp\quad\text{with initial condition}\quad p(t_{i}) = \lim_{t\searrow t_{i}}p(t) - \left(\frac{\partial J_{i}}{\partial y}\frac{\partial h}{\partial x}\right)^T$$
- in practice nearly $O(n_x)$ (2 ODEs and a simple quadrature)
- more involved implementation
- no least-squares optimizers applicable
github.com/amici-dev/amici
high-performance ODE simulation and sensitivity calculation
Uncertainty analysis
Profile likelihoods
- profile likelihoods defined as $\text{PL}_{g(\theta)}(c) = \max_{\theta\in g^{-1}(c)}p(D|\theta)$
- confidence intervals derived from likelihood-ratio tests
- profiles calculated via optimization / integration / local approximation
Bayesian inference
Posterior analysis
Often, we are interested in integrals of the posterior, e.g.
Monte-Carlo sampling can be used to approximate expectations: $$\mathbb{E}(f(\theta)|D) \overset{n\rightarrow\infty}{\approx} \frac{1}{n}\sum_if(\theta_i)$$ with $\theta_i \overset{\text{iid}}{\sim} p(\theta|D)$.
- Posterior mean of a parameter: $\mathbb{E}(\theta|D) = \int \theta p(\theta|D)d\theta$
- Posterior variance (uncertainty): $\mathbb{V}(\theta|D) = \mathbb{E}(\theta^2|D) - \mathbb{E}(\theta|D)^2$
- Credible intervals: $\mathbb{P}(\theta\in[l,u]|D) = \int_l^u p(\theta|D)d\theta$
- Marginals: $p(\theta_1|D) = \int p(\theta|D)d\theta_2\cdots d\theta_n$
Monte-Carlo sampling can be used to approximate expectations: $$\mathbb{E}(f(\theta)|D) \overset{n\rightarrow\infty}{\approx} \frac{1}{n}\sum_if(\theta_i)$$ with $\theta_i \overset{\text{iid}}{\sim} p(\theta|D)$.
How to sample from a distribution?
- Simple distributions: CDF inversion $F_{p(\theta|D)}^{-1}(U[0,1])$
- Rejection sampling: $\theta\sim q(\theta)$, accept if $U[0,1] < p(\theta|D)/(Mq(\theta))$
- Importance sampling: $\mathbb{E}_{p(\theta|D)}(f) = \int f(\theta)\frac{p(\theta|D)}{q(\theta)}q(\theta)d\theta$
does not really scale well to high-dimensional distributions ...
Markov-Chain Monte-Carlo (MCMC)
idea: instead of randomly drawing iid samples, evolve samples via small perturbations to what was already observed
until chain long enough:
- propose $\theta \sim q(\theta|\theta_i)$
- if $U[0,1] < \min\left(1,\frac{p(\theta|D)q(\theta|\theta_i)}{p(\theta_i|D)q(\theta_i|\theta)}\right)$, then $\theta_{i+1}=\theta$, else $\theta_{i+1} = \theta_i$
Markov-Chain Monte-Carlo (MCMC)
- More refined updates (Hamiltonian Monte-Carlo, NUTS, ...)
- Adapt to problem structure
- Parallel Tempering
Further topics
- Outlier control
- Model selection
- Ordinal data
- Mixed effects
- Standards
- Residual analysis
- Structural identifiability
- ...
github.com/icb-dcm/pypesto
parameter estimation toolbox
- optimization, profiles, sampling
- own implementations and interfaces to popular tools
- high-performance computing parallelization
- model selection, visualization, ...
github.com/petab-dev/petab
data format for specifying parameter estimation problems
Literature
- Fröhlich, Fabian, et al. "AMICI: High-Performance Sensitivity Analysis for Large Ordinary Differential Equation Models." arXiv preprint arXiv:2012.09122 (2020).
- Fröhlich, Fabian, Carolin Loos, and Jan Hasenauer. "Scalable inference of ordinary differential equation models of biochemical processes." Gene Regulatory Networks (2019): 385-422.
- Raue, Andreas, et al. "Lessons learned from quantitative dynamical modeling in systems biology." PLoS ONE, 8(9):e74335, 2013.
- Ballnus, Benjamin, et al. "Comprehensive benchmarking of Markov chain Monte Carlo methods for dynamical systems." BMC Systems Biology, 11(63), 2017.
Likelihood-free inference
Likelihood-free Bayesian inference
- common optimization and sampling methods (e.g. MCMC) require the (unnormalized) likelihood
- can happen: numerical evaluation infeasible
- ... but still possible to simulate data $y\sim\pi(y|\theta)$
Example: Modeling tumor growth
based on Jagiella et al., Cell Systems 2017
- cells modeled as interacting stochastic agents, dynamics of extracellular substances by PDEs
- simulate up to 106 cells
- 10s - 1h for one forward simulation
- 7-18 parameters
What we tried |
|
Failed |
Worked |
|
--- |
How to infer parameters for complex stochastic models?
ABC
Approximate Bayesian ComputationRejection ABC
With distance $d$, threshold $\varepsilon>0$,
summary statistics $s$:
until $N$ acceptances:
- sample $\theta\sim g(\theta)$
- simulate data $y\sim\pi(y|\theta)$
- accept $\theta$ if $d(s(y), s(y_\text{obs}))\leq\varepsilon$
A "derivation"
Rejection sampling
Background: Want to sample from $f$, but can only sample
from
$g \gg f$.
Let $f=\pi(\theta|y_\text{obs}), g=\pi(\theta) \Rightarrow \frac{\pi(\theta|y_\text{obs})}{\pi(\theta)} \propto \pi(y_\text{obs}|\theta)$
until $N$ acceptances:
Accepted $\theta$ are independent samples from $f(\theta)$.- sample $\theta\sim g(\theta)$
- accept $\theta$ with probability $\propto \frac{f(\theta)}{g(\theta)}$
Let $f=\pi(\theta|y_\text{obs}), g=\pi(\theta) \Rightarrow \frac{\pi(\theta|y_\text{obs})}{\pi(\theta)} \propto \pi(y_\text{obs}|\theta)$
- not available
- idea: circumvent likelihood evaluation by simulating data and matching them to the observed data
Likelihood-free rejection sampling
until $N$ acceptances:
- sample $\theta\sim \pi(\theta)$
- simulate data $y\sim\pi(y|\theta)$
- accept $\theta$ if $y=y_\text{obs}$
- Acceptance probability: $\mathbb{P}[y_\text{obs}]$
- can be small in particular for continuous data
- idea: accept simulations that are similar to $y_\text{obs}$
Rejection ABC
With distance $d$, threshold $\varepsilon>0$:
until $N$ acceptances:
- sample $\theta\sim \pi(\theta)$
- simulate data $y\sim\pi(y|\theta)$
- accept $\theta$ if $d(y, y_\text{obs})\leq\varepsilon$
- curse of dimensionality: if the data are too high-dimensional, the probability of simulating similar data sets is small
- idea: create an informative lower-dimensional representation via summary statistics
- ideally minimal sufficient statistics
Toy example
$y\sim\mathcal{N}(2(\theta-2)\theta(\theta+2), 1+\theta^2)$,
$y_\text{obs}=2$
$\pi(\theta) = U[-3,3]$
$d=|{\cdot}|_1$
$N=1000$ acceptances
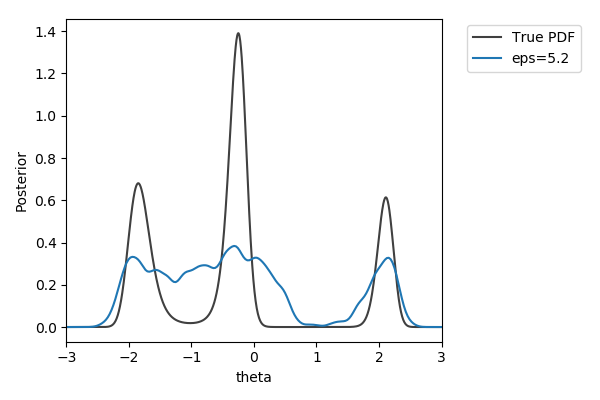
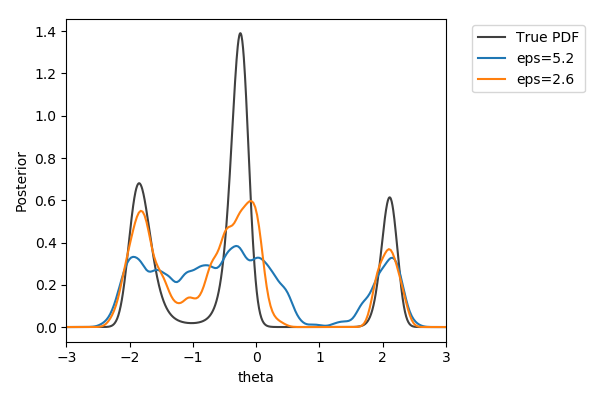

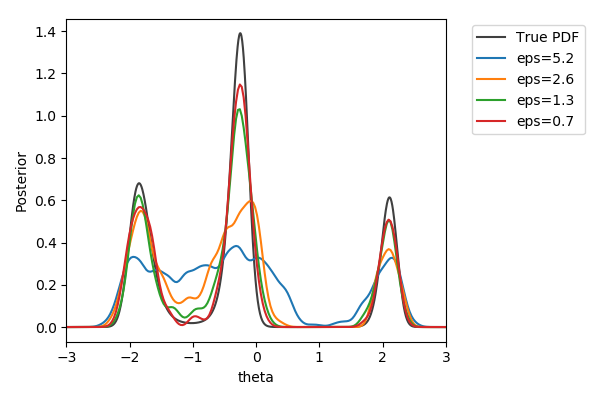

$\pi(\theta) = U[-3,3]$
$d=|{\cdot}|_1$
$N=1000$ acceptances





Approximate Bayesian Posterior
We want: \[\pi(\theta|y_\text{obs}) \propto \color{red}{p(y_\text{obs}|\theta)}\pi(\theta)\]
We get: \[\pi_{ABC}(\theta|s(y_\text{obs})) \propto \color{red}{\int I(\{d(s(y), s(y_\text{obs})) \leq \varepsilon\})p(y|\theta)\operatorname{dy}}\pi(\theta) \approx \frac{1}{N} \sum_{i=1}^N\delta_{\theta^{(i)}}(\theta)\] with distance $d$, threshold $\varepsilon>0$, and summary statistics $s$
Sources of approximation errors in ABC
- model error (as for every model of reality)
- Monte-Carlo error (as for sampling in general)
- summary statistics
- epsilon threshold
Far better an approximate answer to the right question, which is often vague, than an exact answer to the wrong question, which can always be made precise.
John Tukey 1962
Efficient samplers
- Rejection ABC, the basic ABC algorithm, can be inefficient due to repeatedly sampling from the prior
- smaller $\varepsilon$ leads to lower acceptance rates
- many (likelihood-based) algorithms like IS, MCMC, SMC, SA have ABC-fied versions
github.com/icb-dcm/pyabc
Klinger et al., Bioinformatics 2018
Easy to use
# define problem
abc = pyabc.ABCSMC(model, prior, distance)
# pass data
abc.new(database, observation)
# run it
abc.run()
Parallel backends: 1 to 1,000s cores
Parallelization strategies
Klinger et al., CMSB Proceedings 2017Example: Tumor growth model
based on Jagiella et al., Cell Systems 2017Define summary statistics
- 400 cores
- 2 days
- 1.8e6 simulations
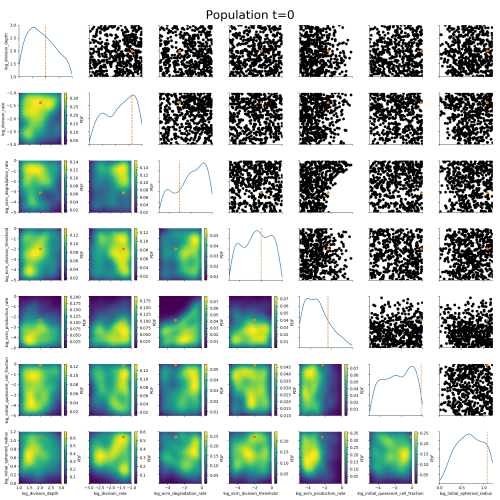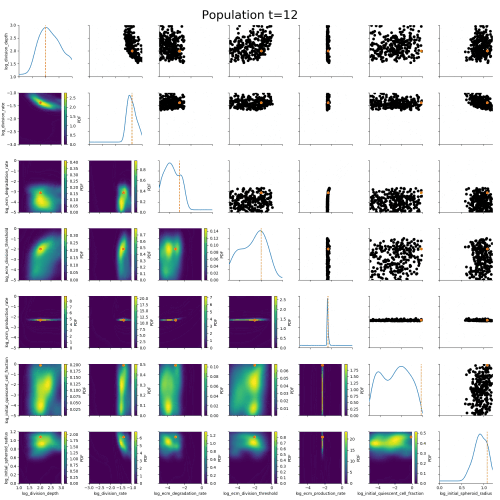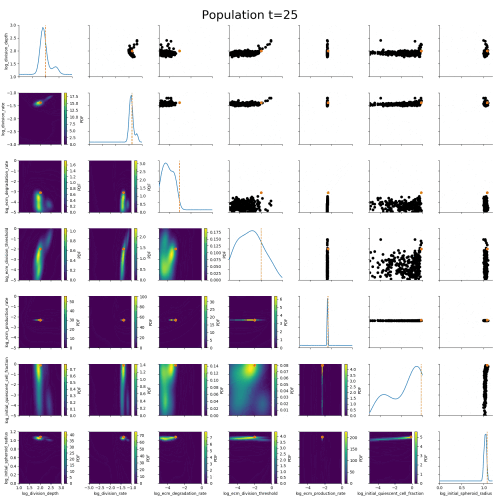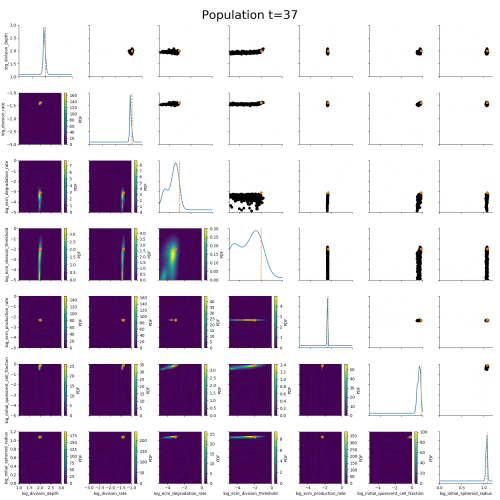
ABC worked where many other methods had failed.
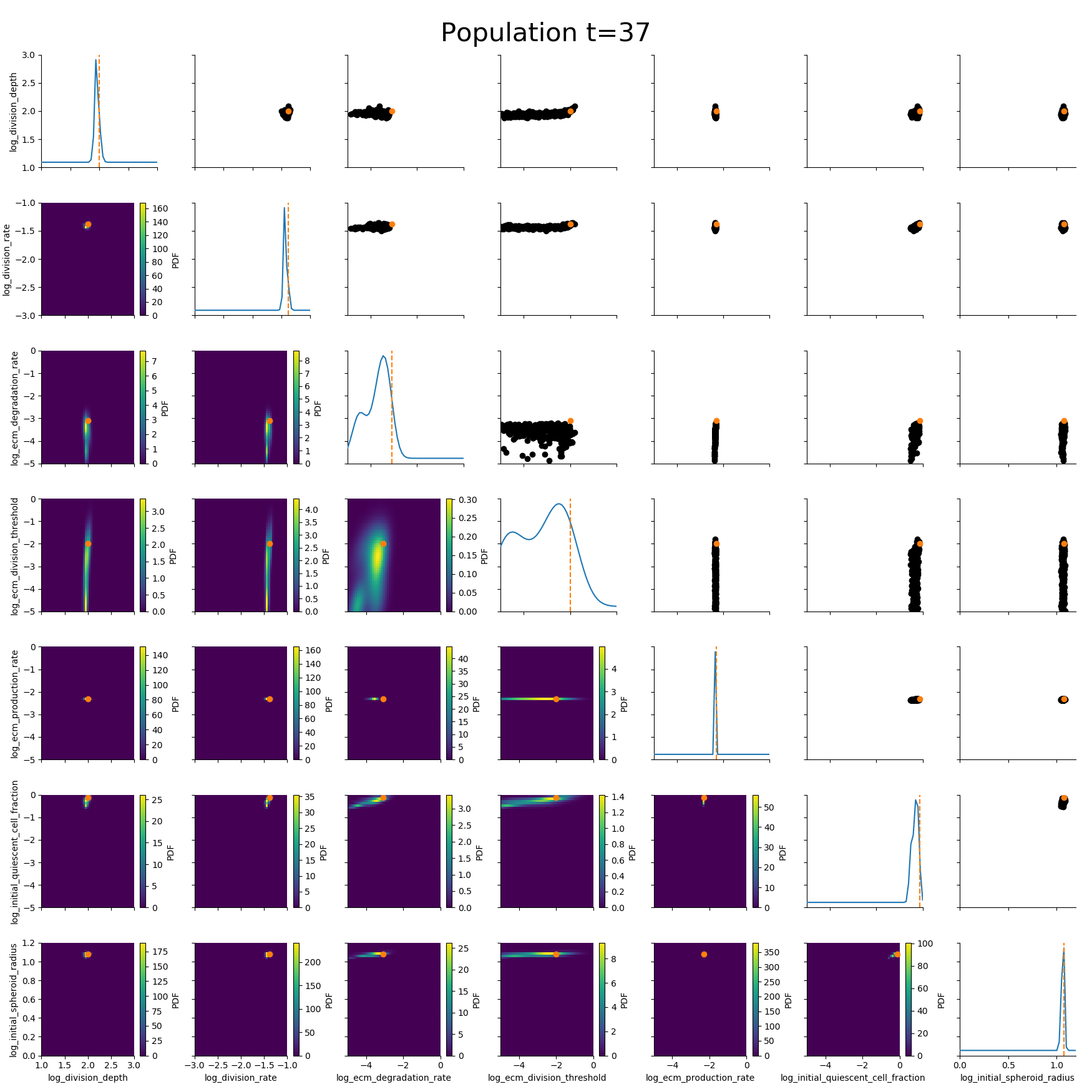
ABC worked where many other methods had failed.
Uncertainty-aware predictions, easy data integration.
Algorithmic details
Adaptive population sizes
Klinger et al., CMSB Proceedings 2017Self-tuning distance functions
based on Prangle, Bayesian Analysis 2015Measurement noise
Schälte et al., Bioinformatics 2020How to efficiently account for measurement noise in ABC?
Further challenges
- Summary statistics calculation
- Epsilon thresholds
- Model selection
- Early rejection
- ...
And ...
...

Joint initiative to perform inference for multi-cellular models
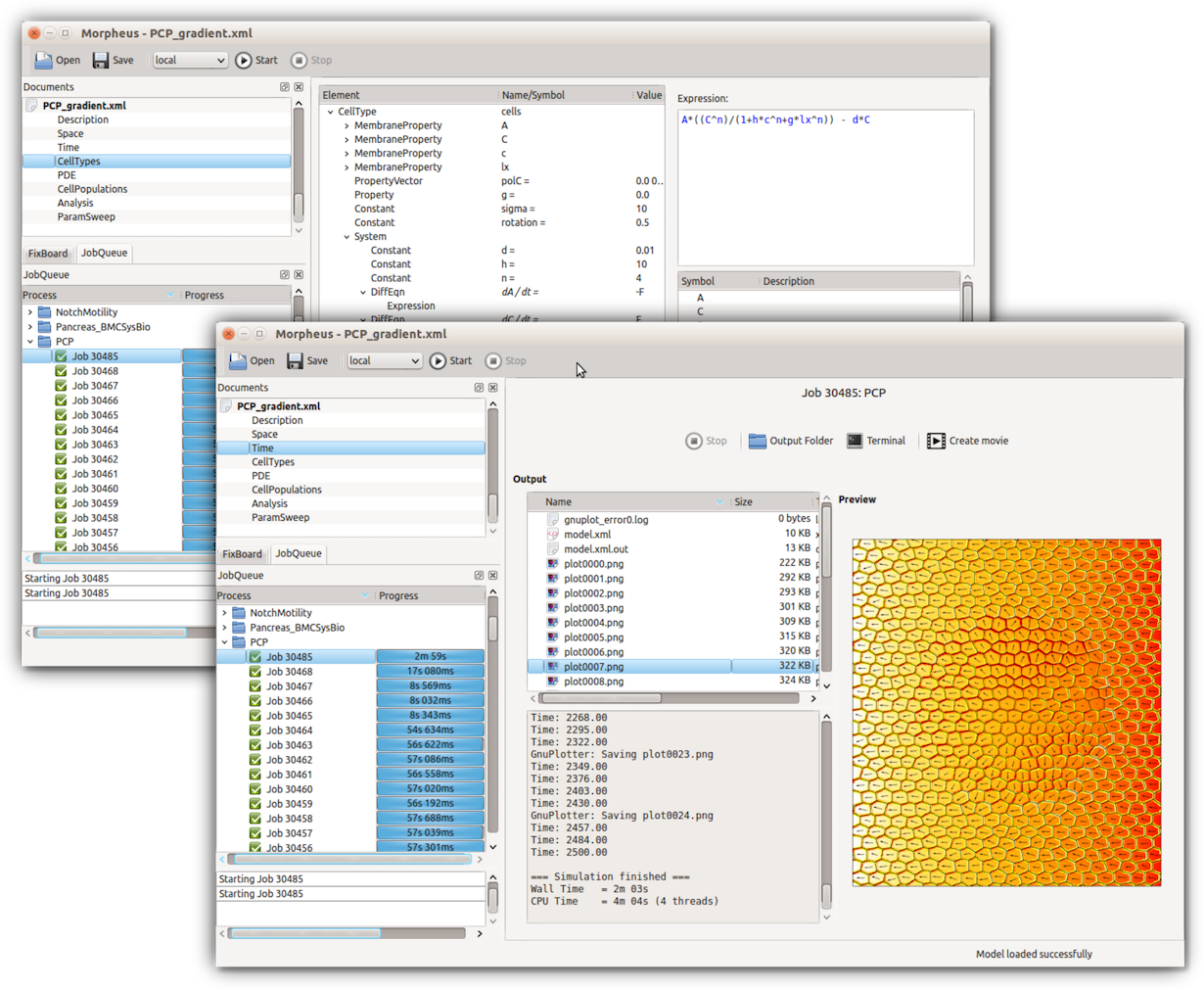
Morpheus toolbox: Staruß et al., Bioinformatics 2014
Literature
- Sisson, Scott A., Yanan Fan, and Mark Beaumont, eds. "Handbook of approximate Bayesian computation." CRC Press, 2018.
- Beaumont, Mark A. "Approximate Bayesian computation in evolution and ecology." Annual review of ecology, evolution, and systematics 41 (2010): 379-406.
- Martin, Gael M., David T. Frazier, and Christian P. Robert. "Computing Bayes: Bayesian Computation from 1763 to the 21st Century." arXiv preprint arXiv:2004.06425 (2020).
Acknowledgments
Thanks to: Jan Hasenauer, Emad Alamoudi, Jakob Vanhoefer, the whole lab, ...